|
|
||||||
| Frank's Home | about DNA and spider silk | Links for learning | A Project: Exploring DNA and silk | Spider anatomy and Learning Center Aids | Web Zoo of Garden Critters | Natasha's Story |
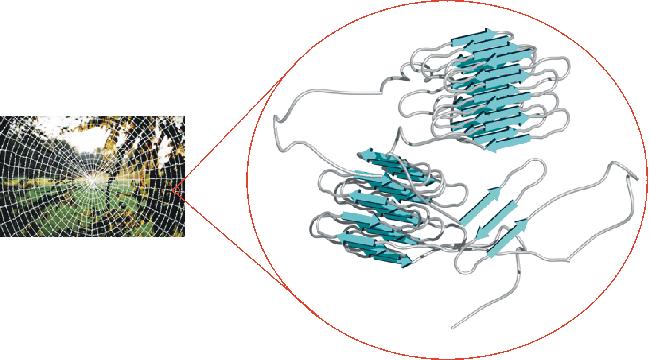
A Science Project for Curious Eyes and an ImaginationI have received several emails from teachers interested in building learning modules around Natasha and her videos. In addition to my photos and video, it might be useful to explore the molecular nature of Natasha and her super-strong silk. Here are some additional additional ideas, based on DNA and protein sequence information that might be useful for personal or classroom use. This figure from Professor Van Hest's group at the Univ. of Nijmegen depicts the molecular nature of spider silk.
This silk is a polymer, contructed by linking together protein moleculare in an array that imparts unbelievable strength to the string of proteins. How does this happen? First, you might start by learning something about the DNA sequence that codes for spidrion, the spider silk protein. Below are links to two amino acid sequences for spidroin, the spider silk, from two different organisms. Can you find any similarities? So what tools do you need? You need the protein (amino acid) sequences and a tool for comparing the two sequences, altering the alignment and telling you what is similar and what is different. So for the sequences and tools, visit the European Bioinformatics Institute and explore their site. (See bioinformatics tool tutorials.) There is a search box at the top of their page where you can look for nucleotide or protein sequences. Try typing spidroin or naphila clavipes in the search box to find interesting sequences.
Next visit their toolbox: Bioinformatics Products and Services and using Align (under sequence analysis) compare the two sequences (see links above). Before you start to do this serious research, first play with a pair of simple sequences so that you discover all the tricks required to get an answer. For example, copy and paste these partial sequences into the Sequence Window (it will open in a separate window of your browser) of their site just to test that you have everything right. Paste sequence_1 into the top sequence window and sequence_2 into the bottom sequence window. Your screen should look something like this.
Now press the run button and wait a bit. You should get a display similar to this. Now for something serious, go to the sequence links above, scroll to the bottom of the page and you'll see a sequence similar to the above. Copy (including the // at the end) and paste it into the sequence window of Emboss (Align). You must add a label at the beginning of each sequence (I used >sequence_1 and >sequence_2). (The sequence format requires you to type ">something" (without the quotes) at the beginning of each sequence (see above) in order to name it.) Now grab the second sequence and paste it in the lower window (remember to include the //). Add a different label at its beginning, then press Run and wait a short time. You have now compared two amino acid sequences for similar regions. This is how new insights are discovered - by looking for patterns where others forgot to look. Its a job for a curious person. Ever wonder about whether something is soluble in water or not? Something is hydrophobic if it has a fear of water. [Did you know that from Greek hydro means water and philos means friend?] You can plot the protein's fear and friendliness for water by pasting a sequence into the window of this site at the Univ of Virginia Now, teach yourself something about what is happening. Its ok to use Google to point you in a good direction. Remember, the goal is to have fun chasing your curiosity. It is fun to do science with the tools that EBI and EMBL. There are more tools at the National Center for Biotechnology Information at NIH . There you will find links to similar databases and tools. Ensembl is another resource for genome browsing . For the super curious, explore the world of trying to predict the structure of a protein from its amino acid sequency. Use this link to the Baylor College of Medicine Protein Secondary Structure Prediction tools or the PredictProtein Server (Columbia University). For the super super curious, explore the 3D structure of membrane proteins |
|||||||||||||